Benchmarking RDF against MDAnalysis#
The algorithms in freud are highly efficient and rely on parallelized C++ code. Below, we show a benchmark of freud.density.RDF and MDAnalysis.analysis.rdf.InterRDF. This benchmark was run on an Intel(R) Core(TM) i7-10750H CPU @ 2.60GHz.
[1]:
import multiprocessing as mp
import timeit
import freud
import gsd
import matplotlib.pyplot as plt
import MDAnalysis
import MDAnalysis.analysis.rdf
import numpy as np
from tqdm import tqdm
[2]:
trajectory_filename = "data/rdf_benchmark.gsd"
r_max = 5
r_min = 0.1
nbins = 75
[3]:
trajectory = MDAnalysis.coordinates.GSD.GSDReader(trajectory_filename)
topology = MDAnalysis.core.topology.Topology(n_atoms=trajectory[0].n_atoms)
u = MDAnalysis.Universe(topology, trajectory_filename, dt=1.0)
rdf = MDAnalysis.analysis.rdf.InterRDF(
g1=u.atoms, g2=u.atoms, nbins=nbins, range=(r_min, r_max)
).run()
[4]:
plt.plot(rdf.results.bins, rdf.results.rdf)
plt.show()
[5]:
freud_rdf = freud.density.RDF(bins=nbins, r_max=r_max, r_min=r_min)
for frame in trajectory:
freud_rdf.compute(system=frame, reset=False)
freud_rdf.plot();
Timing Functions#
[6]:
def time_statement(stmt, repeat=3, number=1, **kwargs):
timer = timeit.Timer(stmt=stmt, globals=kwargs)
times = timer.repeat(repeat, number)
return np.mean(times), np.std(times)
[7]:
def time_mdanalysis_rdf(trajectory_filename, r_max, r_min, nbins):
trajectory = MDAnalysis.coordinates.GSD.GSDReader(trajectory_filename)
frame = trajectory[0]
topology = MDAnalysis.core.topology.Topology(n_atoms=frame.n_atoms)
u = MDAnalysis.Universe(topology, trajectory_filename, dt=1.0)
code = """rdf = MDAnalysis.analysis.rdf.InterRDF(g1=u.atoms, g2=u.atoms, nbins=nbins, range=(r_min, r_max)).run()"""
return time_statement(
code, MDAnalysis=MDAnalysis, u=u, r_max=r_max, r_min=r_min, nbins=nbins
)
[8]:
def time_freud_rdf(trajectory_filename, r_max, r_min, nbins):
trajectory = MDAnalysis.coordinates.GSD.GSDReader(trajectory_filename)
code = """
rdf = freud.density.RDF(bins=nbins, r_max=r_max, r_min=r_min)
for frame in trajectory:
rdf.compute(system=frame, reset=False)"""
return time_statement(
code, freud=freud, trajectory=trajectory, r_max=r_max, r_min=r_min, nbins=nbins
)
[9]:
# Test timing functions
params = dict(
trajectory_filename=trajectory_filename, r_max=r_max, r_min=r_min, nbins=nbins
)
def system_size(trajectory_filename, **kwargs):
with gsd.hoomd.open(params["trajectory_filename"], "rb") as trajectory:
return {
"frames": len(trajectory),
"particles": len(trajectory[0].particles.position),
}
print(system_size(**params))
mdanalysis_rdf_runtime = time_mdanalysis_rdf(**params)
print("MDAnalysis:", mdanalysis_rdf_runtime)
freud_rdf_runtime = time_freud_rdf(**params)
print("freud:", freud_rdf_runtime)
{'frames': 5, 'particles': 15625}
MDAnalysis: (7.632450833324886, 0.16515610292216557)
freud: (0.50654476666629, 0.0329262648249979)
Perform Measurements#
[10]:
def measure_runtime_scaling_r_max(r_maxes, **params):
result_times = []
for r_max in tqdm(r_maxes):
params.update(dict(r_max=r_max))
freud.parallel.set_num_threads(1)
freud_single = time_freud_rdf(**params)
freud.parallel.set_num_threads(0)
result_times.append(
(time_mdanalysis_rdf(**params), freud_single, time_freud_rdf(**params))
)
return np.asarray(result_times)
[11]:
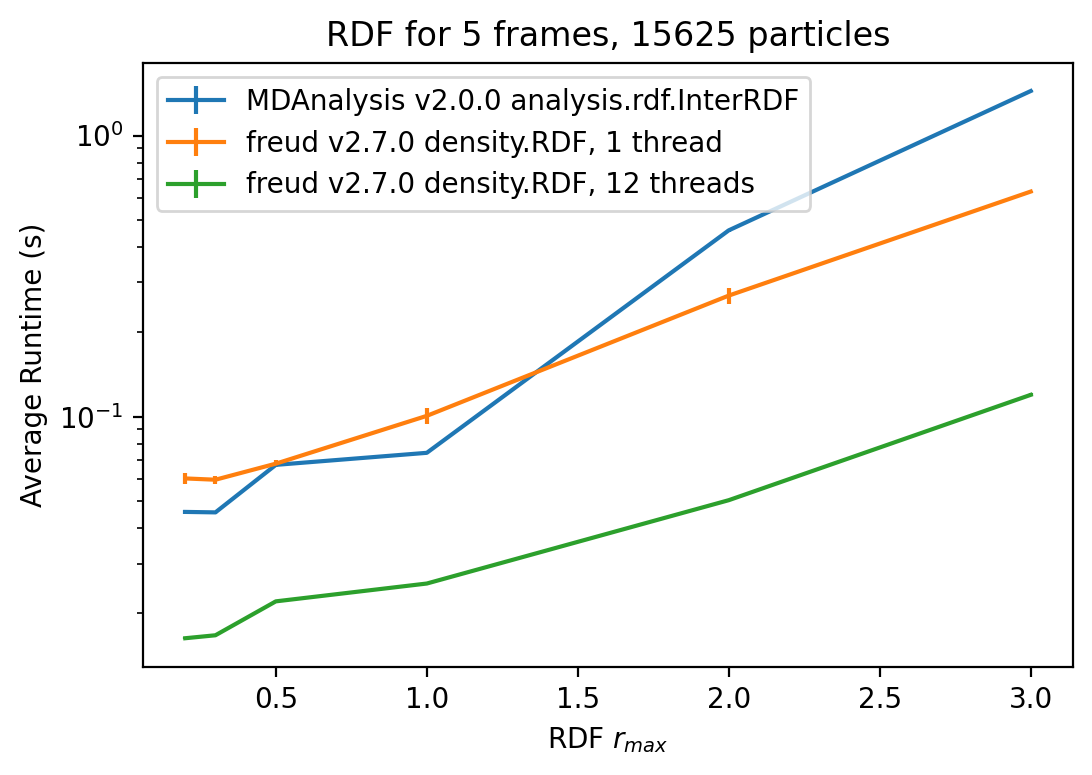
def plot_result_times(result_times, r_maxes, frames, particles):
plt.figure(figsize=(6, 4), dpi=200)
plt.errorbar(
r_maxes,
result_times[:, 0, 0],
result_times[:, 0, 1],
label=f"MDAnalysis v{MDAnalysis.__version__} analysis.rdf.InterRDF",
)
plt.errorbar(
r_maxes,
result_times[:, 1, 0],
result_times[:, 1, 1],
label=f"freud v{freud.__version__} density.RDF, 1 thread",
)
plt.errorbar(
r_maxes,
result_times[:, 2, 0],
result_times[:, 2, 1],
label=f"freud v{freud.__version__} density.RDF, {mp.cpu_count()} threads",
)
plt.title(rf"RDF for {frames} frames, {particles} particles")
plt.xlabel(r"RDF $r_{{max}}$")
plt.ylabel(r"Average Runtime (s)")
plt.yscale("log")
plt.legend()
plt.show()
[12]:
r_maxes = [0.2, 0.3, 0.5, 1, 2, 3]
[13]:
result_times = measure_runtime_scaling_r_max(r_maxes, **params)
plot_result_times(result_times, r_maxes, **system_size(params["trajectory_filename"]))
100%|████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 6/6 [00:10<00:00, 1.79s/it]
[14]:
print(
f"Speedup, parallel freud / serial freud: {np.average(result_times[:, 1, 0] / result_times[:, 2, 0]):.3f}x"
)
print(
f"Speedup, parallel freud / MDAnalysis: {np.average(result_times[:, 0, 0] / result_times[:, 2, 0]):.3f}x"
)
print(
f"Speedup, serial freud / MDAnalysis: {np.average(result_times[:, 0, 0] / result_times[:, 1, 0]):.3f}x"
)
Speedup, parallel freud / serial freud: 4.158x
Speedup, parallel freud / MDAnalysis: 5.450x
Speedup, serial freud / MDAnalysis: 1.207x